In this page, a simple process to use ADAPT-NMR Enhancer on practical ADAPT-NMR outputs will be shown.
First of all, you launch ADAPT-NMR Enhancer, and it will bring up ADAPT-NMR Enhancer program on the screen.
# adapt_nmr_enhancer [ENTER]
Click Open button,
and you will see Open Dialog.
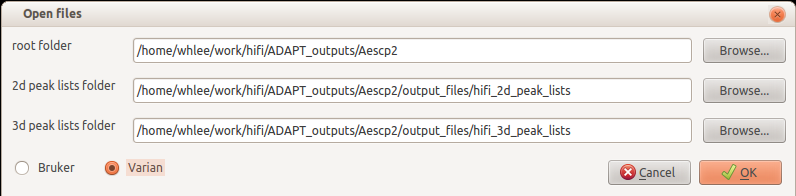
Click Browse button and set your working directory. Don't forget to set the spectrometer vendor you used to collect data by ADAPT-NMR.

When opening is done, set up viewing options for better investigation on quality of peak picking.
For example, this 121 degree tilt plane had a peak picked at a bit shifted place from the likely place. This happens frequently on the shoulder peaks or overlaps. You should focus on peaks with lower priorities. The peaks will be colored darker green in both on the screen and the peak list box.
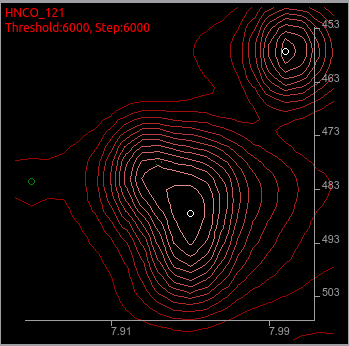
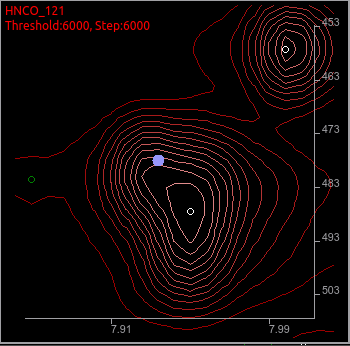
To move it to the more reasonable place, use icons below.

Select a peak (1st icon), delete a peak (4th icon), and add a peak (3rd icon).
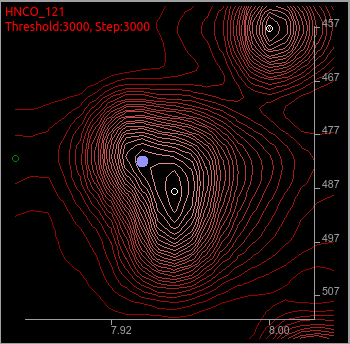
You can see a new peak on the different place from the previous.
Sometimes, you will find a totally noise peak that should be removed without moving.
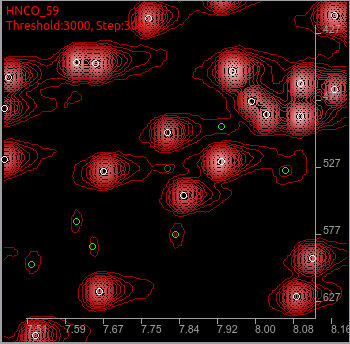
At the center, dark green peak looks a noise. You can judge by using some other convenient tools in the ADAPT-NMR Enhancer to compare between tilt planes, and if you are sure that it is a noise,
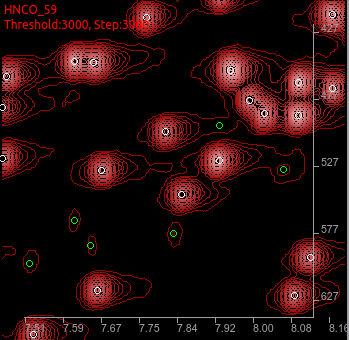
delete a peak to help ADAPT-NMR reiteration.
You should do it throughout experiments. Change the experiment type.
Sometimes, you will find a place where needs a peak that didn't picked.

Add a peak. And if you are very sure about the peak, increase the priority to make it white circle. The line of the circle is now broken, because it is not linked to any 3D constructed peak. However, disregard it at this moment, because re-iteration of ADAPT-NMR will make new constructed peaks and links.

After this work, click Save button.
Then, run ADAPT-NMR.
# $ADAPT_NMR/exe/run_hifi_II.sh $ADAPT_NMR your_working_directory [ENTER]
And, wait for about an hour for ADAPT-NMR to recalculate everything. Then, re-open ADAPT-NMR Enhancer.
# adapt_nmr_enhancer [ENTER]
Click Open, and set working directory just like before.


After re-iteration, you will complete the assignment. First, you will look at Pine Assignment Dialog to find assignments with lower probabilities, or no assignments. If probability of the assignment is low, check the candidates for the atom. Double-click on the candidate will make a selection of the constructed 3D peak which is the nearest to the double-clicked candidate.
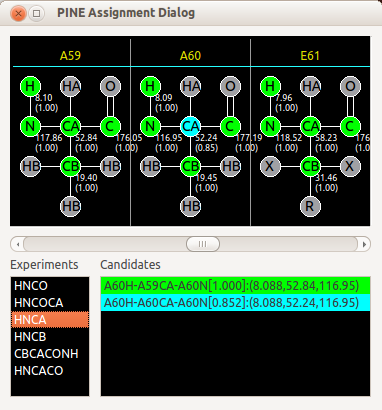
If there is no 3D peak matching with the candidate, ADAPT-NMR Enhancer will ask you whether a new peak on the place of candidates on the spectra.
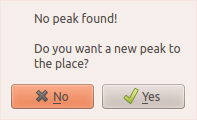
If you choose yes, you can see a 3D peak is added at the bottom of 3D peak list. So, you can see the new peak on the spectra whether the peak makes sense or not.
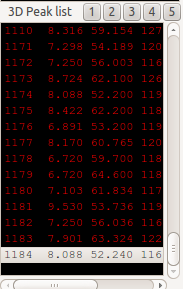
If you are very sure that the new peak, or selected peak is fine and it came from the atoms, you can make an assignment in the Probable Assignment Dialog.
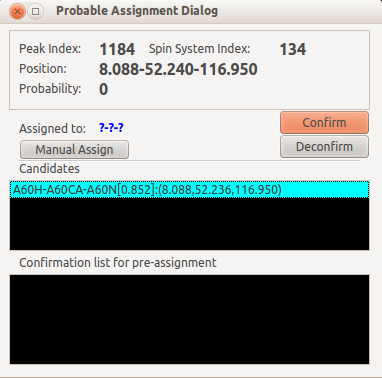
Double click on the candidate and click Confirm button will add the selected candidate into the Confirmation list.
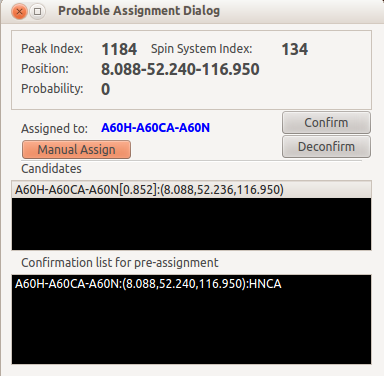
If the peak is not from the candidate list, and you know the assignment, you can click Manual Assign button and make an assignment by yourself.
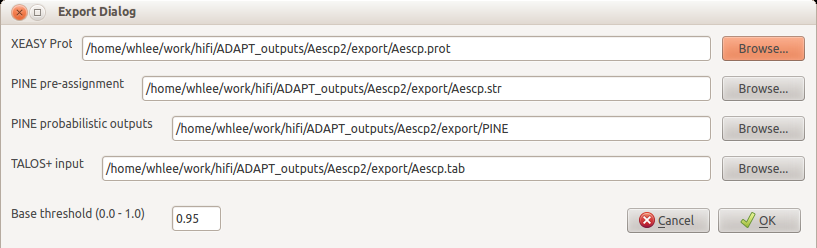
After finishing the assignment, click the export button.

And export the assignment in various formats. You can use the assignment to collect dihedral dstraints by TALOS+, or you can use these data in PINE-SPARKY by using PINE probabilistic outputs. If you have 13C-NOESY, 15N-NOESY data, and you collected sidechains with ADAPT-NMR, you can use XEASY prot to calculate protein 3D structure by using CYANA or PONDEROSA.