PINE does not provide absolute assignments for each peak. It provides possible assignments including the probability that the assignment is correct because PINE uses a probabilistic approach. Multiple possible assignments are converted into labeled objects and the labeled objects are incorporated into SPARKY save files. Since a peak cannot attached to several labels at once in SPARKY, the labels should include position information. The information is saved not only in SPARKY save files but also in newly created files with names that have “_pl” at the tail of the save files because labels are temporary so they can be changed later.
The format of the labels is similar to the following:
possible assignments[probability]:(peak positions)
ex) A14CA-A14N-A14H[0.988]:(60.342,120.231,7.764)
There can be many labels for one peak and once the user determines which assignment is correct, the incorrect labels can be removed. The probable ranks are color coded such that in the default settings, blue labels show the best probability. You can change the color in the PINE2SPARKY program.

2) SPARKY inputs- step 2
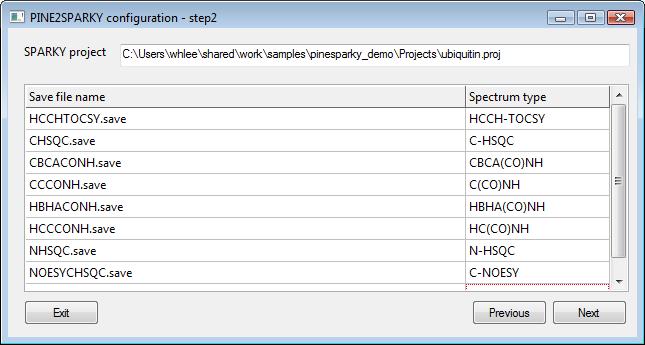
3) Ornaments and Tolerances- step 3
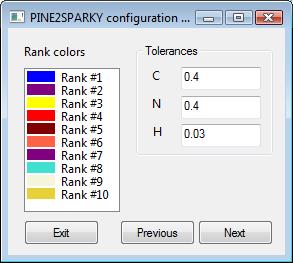
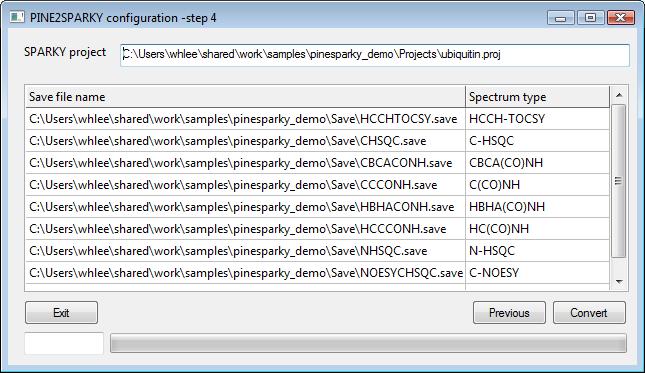
4) SPARKY outputs- step 4
- Old PINE2SPARKY converter (only for MacOSX Tiger)
The user interface of old PINE2SPARKY (only for MacOSX Tiger) consists of roughly four parts: 1) SPARKY inputs, 2) PINE outputs, 3) Sparky outputs, and 4) controls.
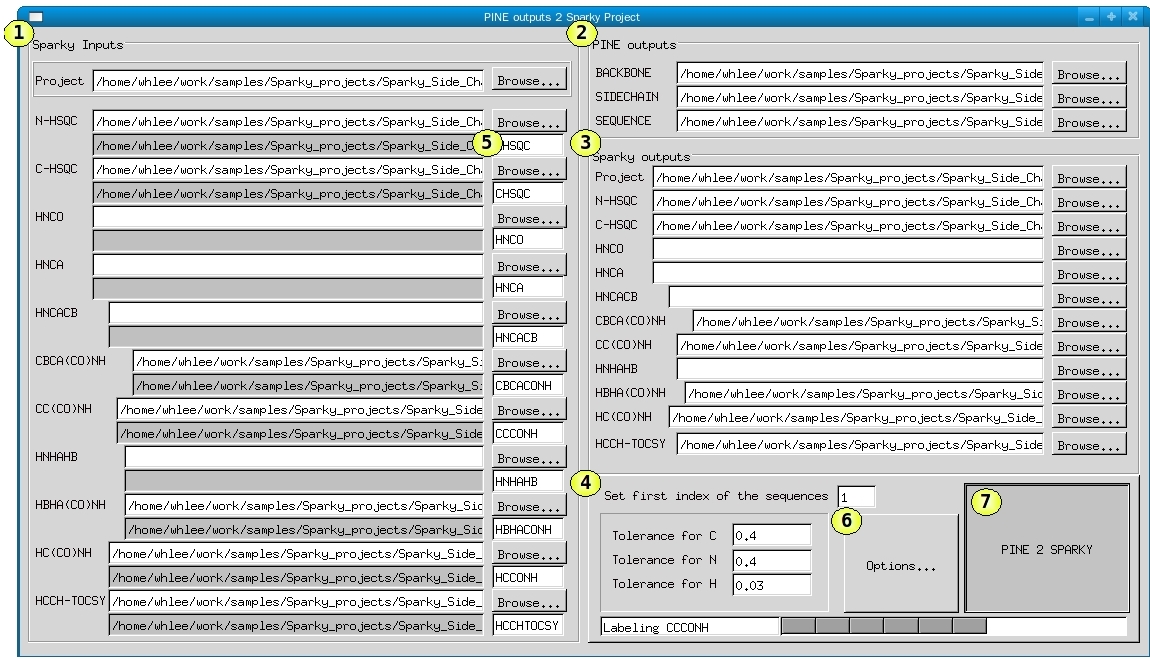
1) Sparky inputs
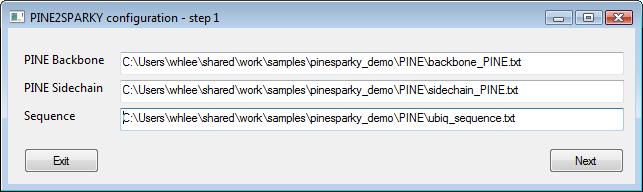
To set the spectra identifier, prepare a peak picked SPARKY project and open it here by pressing “Browse...” button. The save files will be automatically listed in the PINE2SPARKY. If you want to change it, you can press “Browse...” button separately to reset.
2) PINE outputs
You will need a protein sequence file and at least one of the native PINE probabilistic output files. The sequence file can use either the three or one character amino code.
3) Sparky outputs
SPARKY output files are automatically set when you set SPARKY input files. Therefore, performing this program will automatically overwrite your original files. If you do not want to make copies of the original files, you can change the file names by pressing “Browse...” buttons.
4) Controls
(1) Preferences
You can change the default preferences to your own setting. The default for the first index of the sequences is set to 1 and tolerances are set to 0.4, 0.4 and 0.03 for C, N and H-respectively.
(2) Spectrum identifier
PINE2SPARKY generates artificial spin systems for each spectrum and matches these with picked peaks in the SPARKY inputs. So PINE2SPARKY must know spectrum types for SPARKY save files in order to generate proper artificial spin systems. The identifier is used to search for the unique file name in the save files.
(3) Options
The color of the labels for one peak is determined by the ranks of the probabilities. In the default settings, blue is set to represent the highest probability and purple is the next highest. If you want to change colors for ranks, press the “Option...” button and change them.
(4) PINE2SPARKY
After setting up everything you must do, probable assignments from PINE are converted into PINE labels in the Sparky save files by pressing “PINE2SPARKY” button. In addition, “_pl” files which have PINE label information are generated. PINE labels in Sparky save files are not eternal since one can change the position of the labels or remove them entirely, therefore the PINE results should be moved into a permanent file. The time it takes to convert a file is dependent on how many peaks exist in your project.
Prepare input files. 1) Pick peaked SPARKY project and save files that you used as inputs of PINE server. 2) Results from PINE server. They should be native probabilistic outputs. 3) Sequence file – in either three- or one-haracter format.
Fill spectrum identifiers in the sparky input frame. They should be long enough not to be duplicated with names of other spectra.
Click “Browse...” button in the SPARKY input frame and set the SPARKY project file. When you set the project box, the remaining boxes in the sparky input and output frames are filled automatically. If spectrum identifiers are not set correctly, the spectrum will not be filled correctly.
Click “Browse...” buttons in the PINE output frame and set PINE output files. At least one of the backbone and sidechain outputs should be set. Sequence format can be either three letter type or one letter type.
Boxes in the sparky output frame are exactly same as boxes in the sparky input frame. If you do not want to overwrite to your original SPARKY project and save files, you can change target file names by clicking “Browse...” and setting other names. Instead of changing every box, we recommend you to have a complete copy of sources not to lose your data.
Set preferences. First index of sequence index is set to 1 for default. Tolerances are set to 0.4, 0.4 and 0.03 for C, N and H. They are changeable. Click “Options...” button if you want to change probability rank colors.
Finally, click “PINE2SPARKY” button. It will take a minute to finish.